

|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
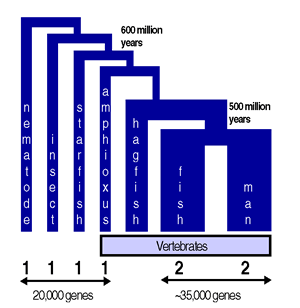
"Essentially all genes and pathways shown to be important in cell-, developmental-
and disease-biology have been found to be conserved between worm and man.
This conservation applies to the number and type of protein families, gene structure,
the hierarchy of genes in genetic pathways and even gene regulation.
A consequence of this conservation is that human genes can readily be inserted into the worm genome, to functionally replace the worm genes even in complex cell biological and signal transduction pathways. Conversely, key worm genes identified using genetics can be used to trigger specific biochemical processes in human cells." *(Graph and quote taken by permission from the Devgene website cited below) |